Joint article by Oxford PharmaGenesis, Digital Science and Pfizer reaches most reads in Cancer Informatics over the last 6 months
03 October 2022
Share this page
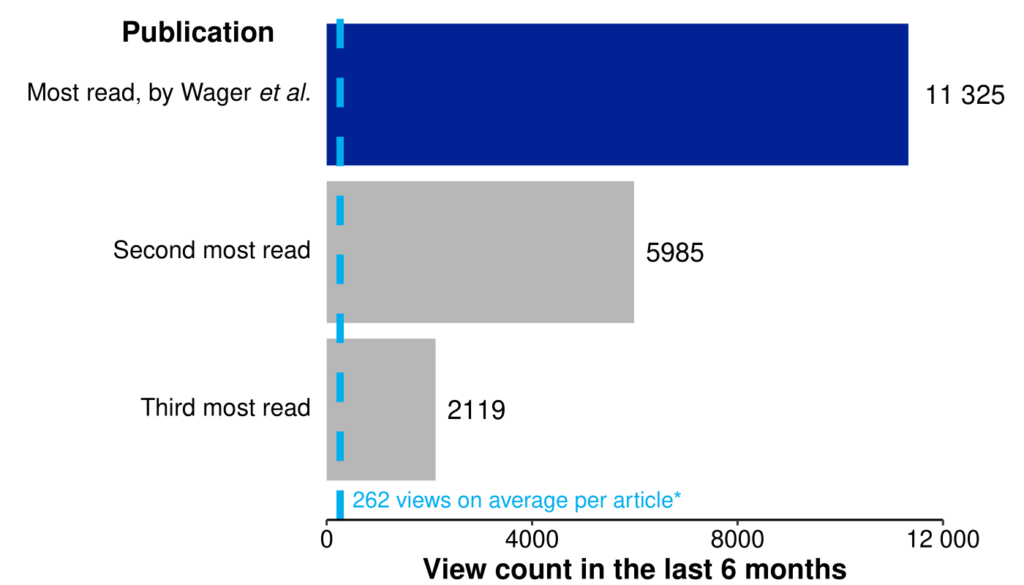
Published in late March in Cancer Informatics, our paper (Wager et al., 2022) has accumulated 11 000 views in the last 6 months. This is twice as many as the second most read article in the journal, five times that of the third most read, and 43 times the number of views that the average article in Cancer Informatics received in 2021. This remarkable achievement crowns a 2-year collaboration between Pfizer, Digital Science and Oxford PharmaGenesis.
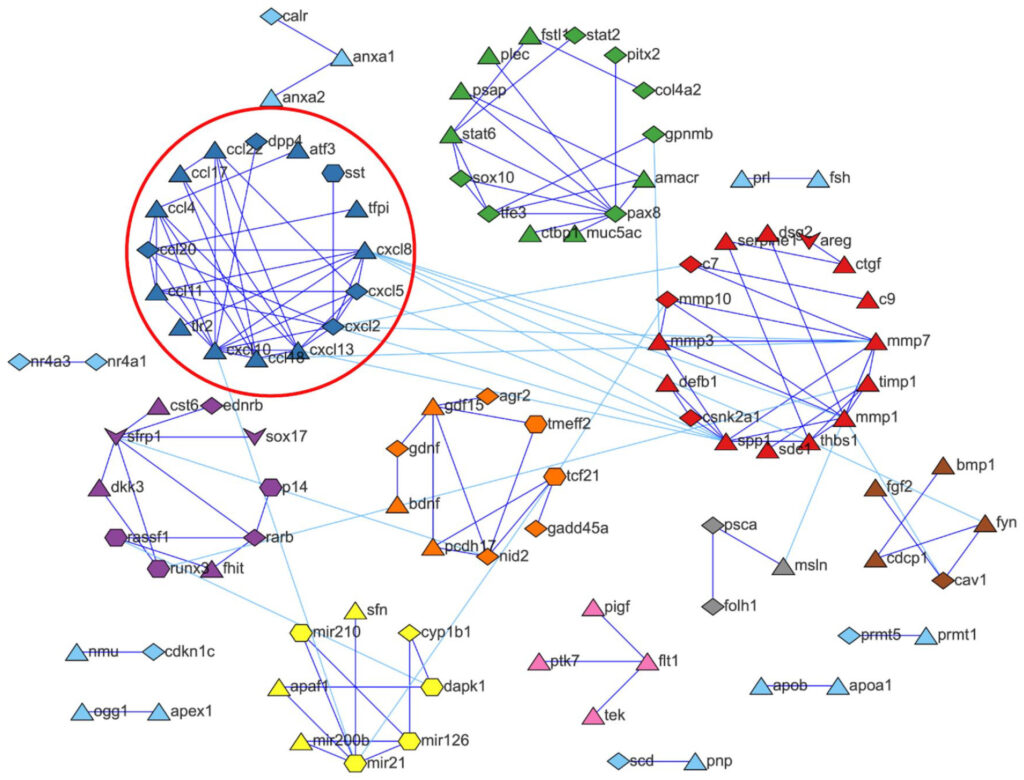
The publication presented a novel automated literature analysis method that leveraged natural language processing to identify emerging cancer biomarkers. Relationships between these biomarkers were also revealed, and some had not yet been described in existing protein interaction databases. Although the method specifically identified cancer biomarkers, it can be adapted to identify relevant literature for other diseases that may be overlooked by keyword searches.
A validated method to provide biological insights
The method identified biomarkers that frequently appeared close to oncology terms within publications, and biomarkers that were in proximity of each other were marked as being potentially associated biologically. To focus on biomarkers with emerging research interest, the method further selected clusters of biomarkers with a high publication growth rate; that is, an increasing number of publications between 2015 and 2020. Finally, in order to validate the method, the authors manually reviewed the publications that contained the selected biomarkers, which confirmed the biological relevance of the output.
Success made possible by a multidisciplinary team
Although the method was validated by biomedical experts, it overcame computational challenges, such as the identification of synonyms (including abbreviations and acronyms), the distinction between homonyms, and the recognition of several biomarker categories (such as genes and proteins). Furthermore, access to full-text publications and data processing was facilitated by Orion Penner and Bob Schijvenaars at Digital Science.
These technical achievements only matter if they can help to meet a scientific objective. One such objective was provided by our Pfizer clients, Dheepa Chari, Global Lead of Oncology Scientific Communications, and Steffan Ho, Vice President and Head of Translation Oncology, who recognized the opportunity to use literature analysis in support of cancer research.
Finally, the popularity of the publication highlights the expertise of Oxford PharmaGenesis, who have effectively communicated research that is at the intersection of biomedical and data science. This was only possible through effective teamwork between internal collaborators: author and writer Kim Wager, author Tomas Rees, editor Allie Gonzato, and personal assistant Emma Mackey.
Click here to read the full paper.
